Blog
30 September 2025

Redefining Translation Studies: The Power of Modern Ribosome Profiling
Discover how ribosome profiling is transforming translatomics, from its origins and cutting-edge methodologies to advanced bioinformatics and breakthrough applications in cancer, neurodegeneration, infectious diseases, and more.
Why does Ribosome profiling matter now?
Ribosome profiling is transforming RNA research by providing high-resolution insights into translation, the crucial step between gene expression and protein synthesis. Unlike transcriptomics or proteomics alone, Ribo-Seq captures which mRNAs are actively translated in real time, offering unmatched visibility into translational dynamics under normal and disease conditions. This advanced RNA sequencing technique is now essential for biotech and pharmaceutical companies focused on RNA-based drug development, as well as academic labs studying gene regulation, cancer biology, and neurodegeneration. With recent improvements in Ribo-Seq protocols and bioinformatics, researchers can now decode translational control mechanisms with unprecedented precision, making ribosome profiling a critical technology for modern molecular biology and therapeutic discovery.
What is Ribosome profiling?
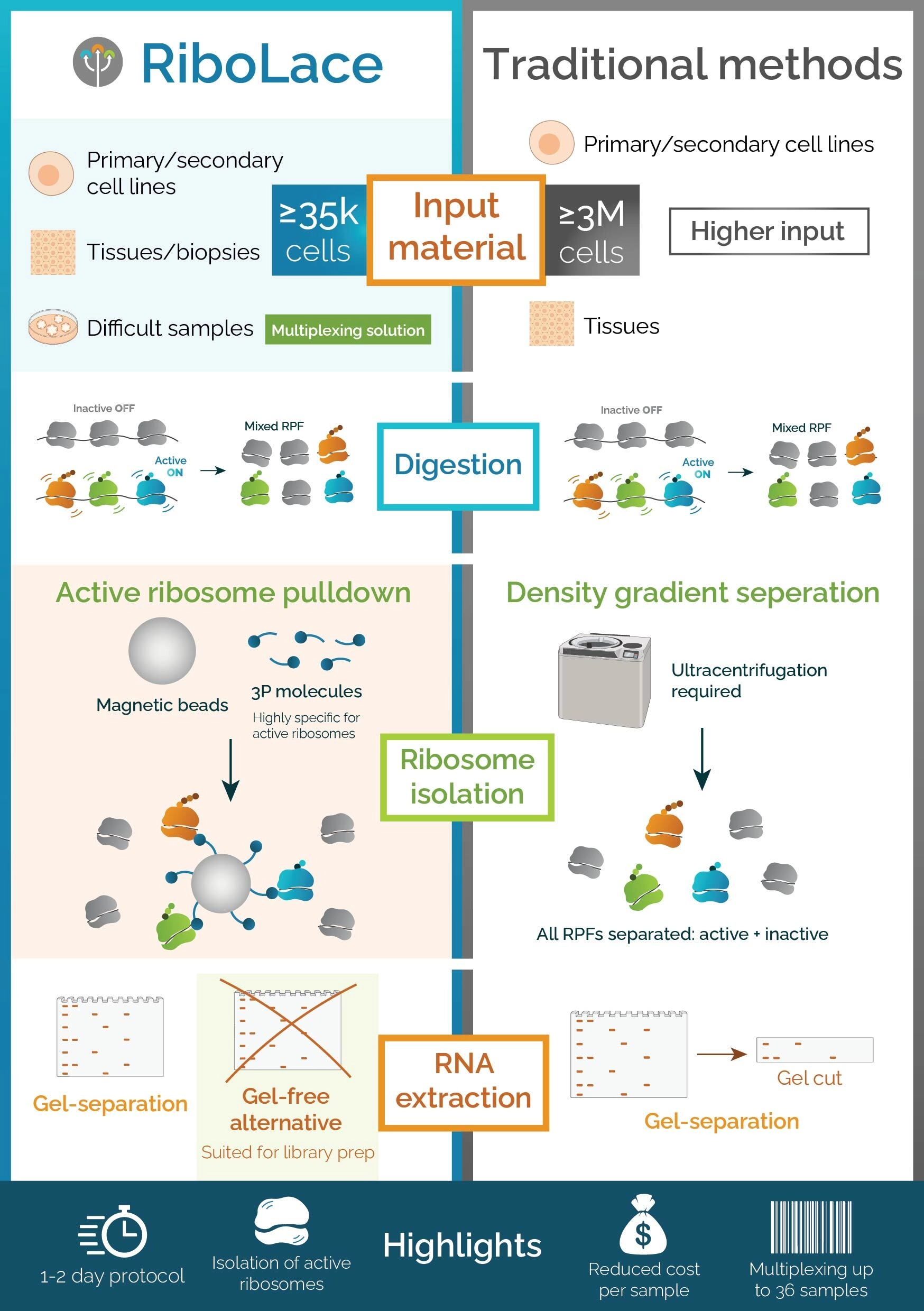
Ribosome profiling (Ribo-Seq) is based on the idea that translating ribosomes protect short mRNA fragments from nuclease digestion. Practically, living cells are treated with a translation inhibitor (e.g., cycloheximide) or flash-frozen instantly to arrest translation. Following cell lysis, an RNase or a tailored nuclease cocktail removes exposed mRNA, preserving ribosome‑protected fragments (RPFs), ~28 nucleotides in length, in eukaryotic species. These RPFs are purified from isolated ribosomes, historically via sucrose-gradient ultracentrifugation and gel excision, with modern-day alternatives including gel-free affinity capture methods to reduce sample loss. After adapter ligation, reverse transcription, and PCR amplification, the library of RFPs is subjected to Next Generation Sequencing (NGS). Finally, reads are aligned to the reference genome/transcriptome, generating a high-resolution map that pinpoints ribosome positions at single-codon resolution.
Ribo-Seq reveals translational control, uncovering non-canonical open reading frame (ORF) activity, e.g., upstream or downstream ORF; codon-specific pausing, and condition-dependent shifts in translation that are unattainable by RNA-Seq or proteomics.
History and Development
Ribosome profiling emerged from early polysome profiling studies that sorted mRNAs by ribosome load using a sucrose gradient but lacked positional resolution. The breakthrough came in 2009 when Ingolia et al. introduced the use of nuclease to isolate individual ribosomes and their RPFs. They then used deep sequencing to map RPFs genome-wide in yeast, revealing non-AUG initiation and codon-specific pausing under stress. This unveiled Ribo-Seq as a precise tool for in vivo translation analysis.
Over the years, Ribo‑Seq was optimized for bacteria, mammalian cells, and plants, with improvements in:
- Lysis buffer to preserve ribosomes.
- Nuclease digestion for optimal RPF length.
- Gel-free purification.
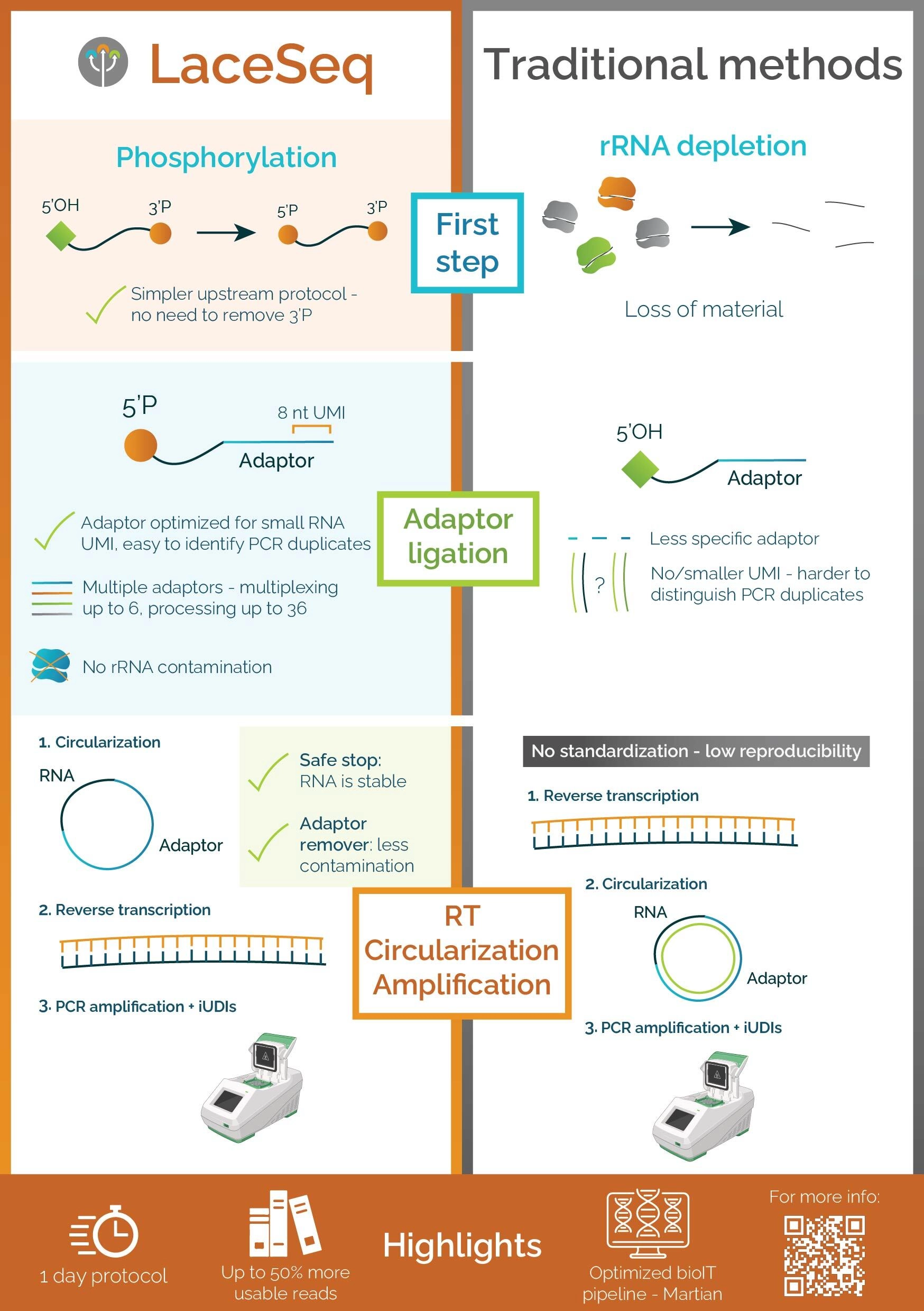
By 2015, Ribo-Seq had evolved into a reproducible, nucleotide-resolution protocol essential for studying translational control in health and disease. Immagina's Ribolace, combined with the downstream Laceseqworkflow (Fig. 1), offers a cutting-edge solution for comprehensive analysis of actively translating ribosomes. RiboLace utilizes a puromycin-derived capture molecule to selectively isolate elongating ribosomes, allowing for the precise recovery of ribosome-protected mRNA fragments without the need for gel purification. Following ribosome isolation, the LaceSeq protocol enables library preparation optimized for next-generation sequencing, preserving translational context while minimizing bias and sample loss. Together, RiboLace and LaceSeq provide an integrated, gel-free, and highly reproducible platform to study translation dynamics, translational control mechanisms, and ribosome-mRNA interactions across diverse biological systems.
Bioinformatic analysis: from data to discovery
Bioinformatic analysis is essential for interpreting Ribo-Seq data, turning raw sequencing reads into insights about translational regulation. The process begins with adapter trimming, quality filtering, and removal of ribosomal RNA reads, followed by alignment to the genome or transcriptome. Once mapped, ribosome-protected fragments are analyzed to assess key quality metrics such as fragment length distribution and triplet periodicity, confirming the data’s translational relevance. Downstream, quantification of ribosome occupancy allows the identification of Differentially Translated Genes (DTGs) across experimental conditions. When combined with RNA-Seq data, this enables the calculation of Translation Efficiency (TE) a robust metric that uncovers changes in protein synthesis independent of mRNA abundance. Additional analyses, such as open reading frame (ORF) mapping and periodicity-based detection of translation events, further enhance the resolution of the data, offering a comprehensive view of gene expression regulation at the translational level.
Applications across biomedical sectors
The power of ribosome profiling lies in its wide-ranging applications across biology and medicine:
Oncology: Ribo-seq has revealed altered translational control of oncogenes, tumor suppressors, and non-coding regions previously thought to be untranslated. Notably, Ribo-Seq has uncovered the translation of small open reading frames (sORFs) and uORFs that modulate key oncogenic pathways, adding a new layer of complexity to tumor biology and potential therapeutic targeting Zang et al., 2025.
Neurodegeneration: Neurons are uniquely vulnerable to disruptions in proteostasis. In Alzheimer’s and Parkinson’s models, Ribo-Seq uncovers aberrant translation of aggregation-prone peptides and stress-induced uORFs, with region-specific defects observed in post-mortem brain tissue.
Infectious Disease: Viruses must co-opt host translation while evading innate immunity. Ribo-Seq maps viral and host translation simultaneously, exposing mechanisms like IRES initiation and leaky scanning helpful for developing targeted antivirals.
Developmental Biology: In early embryos, Ribo-Seq tracks translational waves during the maternal-to-zygotic transition, showing how selective translation precedes and guides transcriptional activation and cell fate.
Metabolic Disorders and Immunology: Under stress or inflammation, Ribo-Seq identifies selective translation of unfolded protein response (UPR) regulators in liver cells and finely tuned cytokine translation in activated T cells, advancing insights into metabolic disease and immune response.
Challenges and Solutions in Ribosome Profiling
Ribosome profiling (Ribo-Seq) has revolutionized our understanding of translational control, but its implementation is not without significant technical and biological challenges. One major hurdle is the complexity and variability of sample preparation, particularly the requirement for precise timing and rapid lysis to preserve ribosome-mRNA complexes. Traditional protocols rely heavily on sucrose gradient ultracentrifugation and gel-based RNA purification, which are labor-intensive, prone to user variability, and limited in scalability.
Another key challenge lies in the interpretation of Ribo-Seq data. The presence of ribosome-protected fragments (RPFs) does not always equate to productive translation, as paused ribosomes or stalled complexes can confound signal interpretation. Furthermore, ribosome density alone may not reflect translational efficiency or protein output, especially in complex biological contexts like cancer or stress responses.
Solutions to these limitations are now emerging. Technologies such as RiboLace, developed by Immagina, offer a gel-free approach that selectively captures elongating ribosomes using a puromycin analog. This reduces hands-on time, improves reproducibility, and avoids contamination from non-translating ribosomal subunits. When combined with LaceSeq, an optimized library preparation protocol, researchers gain access to high-quality, strand-specific sequencing libraries suitable for precise quantification and positional mapping of translation events. Additionally, advancements in computational pipelines now incorporate metagene analyses, triplet periodicity checks, and machine learning models to distinguish active translation from noise, further improving data interpretation. By integrating these technological advancements, ribosome profiling is becoming more accessible, scalable, and biologically informative, paving the way for deeper insights into translational regulation across development, disease, and therapeutic discovery.
Want to see how our Immagina's solutions help scientists study RNA in new ways? Take a look at the technologies.
Conclusion and Future Trends in Ribo-seq
Ribosome profiling transforms our capacity to decode the translatome. The next wave of innovation in Ribo-seq is focused on enhancing sensitivity and spatial resolution. Techniques like nano-scale Ribo-Seq now enable translational insights from nanogram-level inputs, opening new avenues for studying rare cell populations, micro-dissected tissues, and uncovering short open reading frames (sORFs) that encode regulatory micropeptides. Parallel advancements in single-cell translatomics are beginning to map translation at subcellular resolution, capturing how translational programs shift in specific microenvironments, developmental stages, or disease states. These tools are helping us understand how neuronal subtypes adjust translation during aging or proteotoxic stress, and how spatially localized translation contributes to processes like organogenesis. Looking ahead, future iterations may bring real-time, even live-cell capabilities, positioning ribosome profiling as a cornerstone in both precision diagnostics and RNA-targeted therapeutics, particularly in oncology, neuroscience, and developmental biology.
FAQs
Q: How does ribosome profiling differ from polysome profiling?
Polysome profiling provides a global overview of translation by separating and analyzing free mRNAs, monosomes, and polysomes, offering insight into the number of ribosomes associated with each transcript. Ribosome profiling, in contrast, is more precise and offers nucleotide-level resolution of ribosome positions on specific mRNAs. While both techniques reveal translation dynamics, ribosome profiling can dissect the fine details of initiation, elongation, and termination events.
Q: What are the main applications of ribosome profiling? Ribosome profiling helps scientists see which proteins are being made in a cell at any given time. It is used to study how genes are turned into proteins, how cells respond to stress or drugs, and how translation changes in diseases like cancer. Researchers use it to find new drug targets, understand disease mechanisms, and improve RNA-based therapies.
Q: Why should I do Ribo-Seq over/in addition to RNA-Seq? RNA-Seq shows which RNAs are present in a sample, but it doesn’t tell you if they are being translated into proteins. Ribo-Seq goes one step further by showing which RNAs are actively used by ribosomes to make proteins. Using both methods together gives a clearer picture of gene expression, helping you understand not just what’s written in the RNA, but what’s actually being made into functional proteins. Q: My RNA-Seq data does not align with the proteomics data. Can I use Ribo-Seq to bridge this gap? Yes, Ribo-Seq can bridge the gap by showing which mRNAs are actively translated, unlike RNA-Seq, which only shows expression. It helps explain why some proteins don’t match RNA levels by capturing real-time translation activity.
Q: What kits are recommended?
It depends on your needs. Traditional ribosome profiling protocols (‘gel-cut’ or Ingolia method) yield data from clean ribosome-protected fragments (RPFs) but are labor-intensive and require equipment such as ultracentrifuges. Immagina’s ALL-IN-ONE RiboLace Gel Free kit instead uses magnetic bead–based RiboLace technology to capture actively translating ribosomes, removing the need for gel purification and size-selection. This streamlines the workflow, reduces variability, and produces high-quality data comparable to gel-cut approaches. Read more in our application note.
Q: Can Ribo Seq detect micropeptides?
Yes, usually from short ORFs (20–100 amino acids) that are often missed by standard proteomics.