nano-tRNAseq
Studying the function of tRNAs in protein translation is fundamental for understanding their role in several cellular processes that can be impaired in various diseases. Immagina nano-tRNAseq is the only technology that simultaneously quantifies full-length, native-state tRNAs and detects chemical modifications in a single experiment.
Service overview
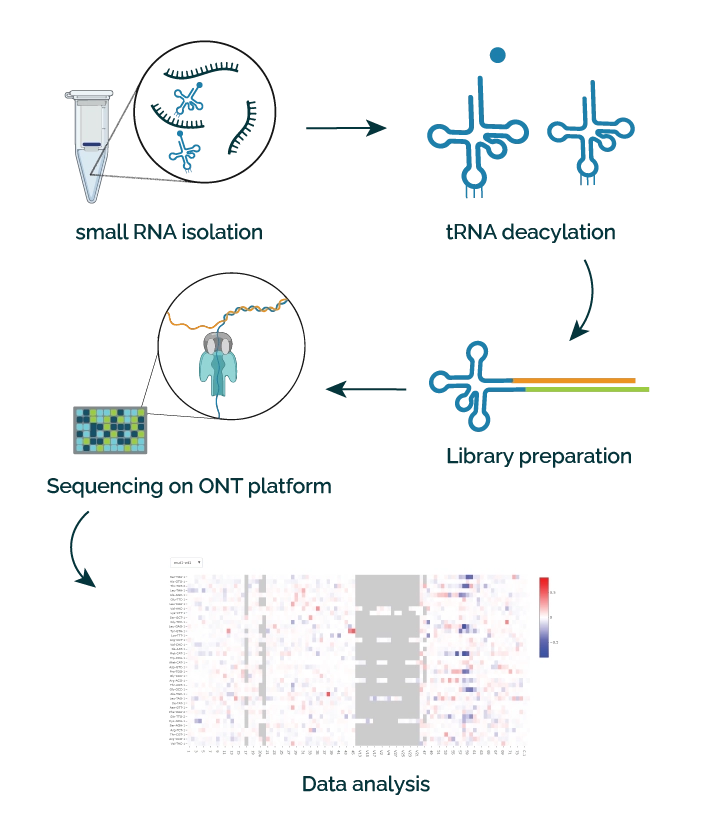
Our service builds upon the pioneering work at the laboratory of Dr. Eva Novoa (Centre for Genomic Regulation (CRG), Barcelona Institute of Science and Technology, Barcelona, Spain. Their developments harness Oxford Nanopore Technologies (ONT) sequencing systems to directly sequence tRNA, bypassing the need for cDNA/PCR sequencing.1 With the ONT technology, as a tRNA molecule traverses a protein nanopore, each base generates a unique electrical current, converted into the tRNA sequence. Direct nano-tRNA sequencing comprises the following steps:
- Small RNA isolation: enriched from total RNA
- tRNA preparation: deacylation, which allows for high-efficiency, specific tRNA-adaptor binding
- Library preparation: allows for multiplexing of up to 6 samples
- Sequencing on ONT platform
- Data analysis: improved base-calling for short RNA sequences.
Technical applications
- Expand translatome studies
- Quantify tRNA abundance
- Detection of tRNA modifications
- Explore ribo-embedded tRNA sequencing
Delivery time: 4 to 6 weeks
Service description and outcomes
- Pre-service consultation to establish your experimental design
- Input material: total RNA extracted from all living organisms (nano-tRNA seq method enables low sample input)
- Complete and interactive bioinformatics report
- Access to secure data transfer portal
- Multiplexing up to 6 samples
- High-efficiency data processing

