Polysome Profiling
Polysome profiling is an established technique for understanding how efficiently genes are translated into proteins. Our Polysome profiling service offers a powerful tool for deciphering how cells function in health and disease.
Service overview
Polysome profiling separates the components of a cell lysate by ultracentrifugation on a sucrose gradient according to their sedimentation coefficient. The main purpose of this separation is to discriminate how many ribosomes are loaded on each mRNA to assess the translational level. More ribosomes bound to an mRNA drive a faster sedimentation rate during gradient centrifugation.
How it works
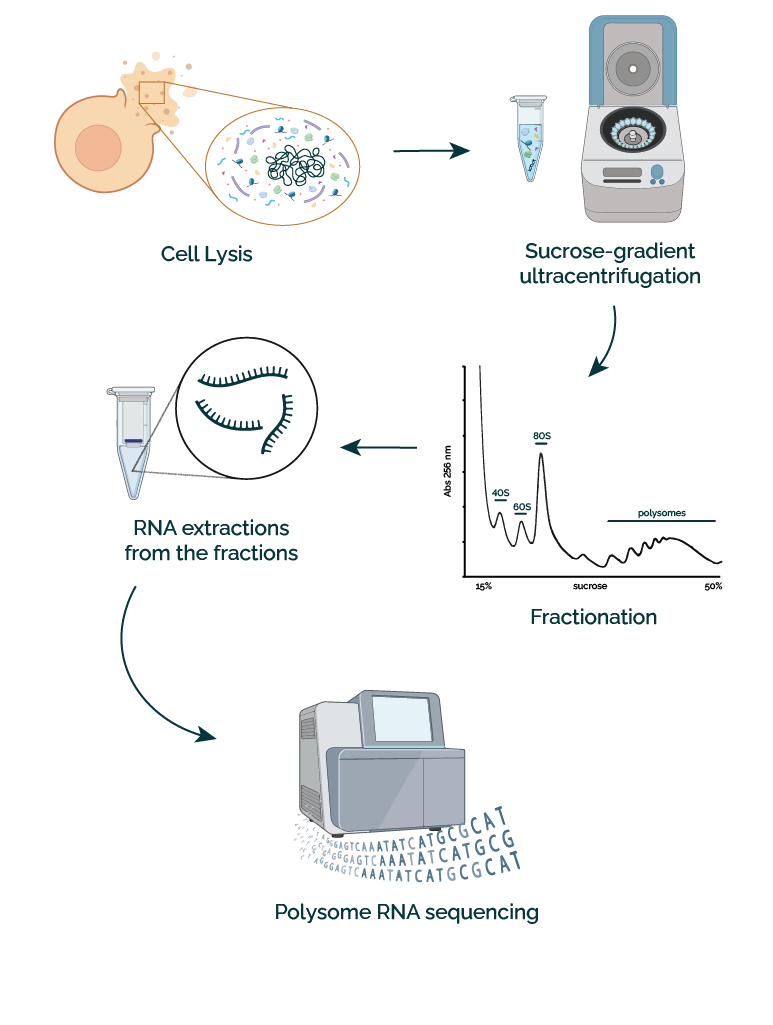
- Cell lysis: The procedure begins with cell lysis. The resulting lysates contain free mRNAs, small and large ribosome subunits (40S and 60S respectively, in eukaryotes), monosomes, and polysomes with associated transcripts.
- Sucrose-gradient centrifugation: The lysates are loaded on a linear sucrose density gradient and ultracentrifuged to separate lysate components.
- Fractionation: After ultracentrifugation, optical density is used to differentiate the gradients, which are then separated and collected into fractions.
- RNA extraction from the fractions: subpolysomal fractions (non-translated mRNAs) and polysomal fractions (translated mRNAs) are then processed for RNA extraction.
- Polysome RNA sequencing: subpolysomal and polysomal extracted RNAs are converted into cDNA libraries and sequenced.
Technical applications
- Studying translational control
- Identification of new mechanisms of translational regulation
- Quantifying transcript translational efficiency
- Translation of alternative splice isoforms
- Study of ribosomal-associated RNAs
Delivery time: 10 to 12 weeks
Service description and outcomes
- Pre-service consultation to establish your experimental design
- Input material: flash-frozen cell pellets or tissues
- Sucrose gradient absorbance profiles (calculation of fraction of ribosomes on polysome)
- RNA extraction and library preparation
- NGS sequencing
- Bioinformatic analysis

