Active Ribosome Profiling
Unveiling the Actively Translated Population
How it compares to standard Ribo-Seq
- A population of messenger RNA molecules exists in a "stalled" state. In this scenario, ribosomes are loaded onto the mRNA and ready for translation, but actual protein production does not occur.
- Standard Ribo-Seq captures all ribosomes on mRNA, including those paused and not actively translating proteins. This underestimates true translation efficiency and weakens the connection to protein levels measured by proteomics methodologies.
- Active Ribo-Seq only targets actively translating ribosomes. This allows the discrimination of the active and the stalled ribosome populations and offers a more accurate picture of translation efficiency.
How it works
- Conventional Ribo-Seq techniques rely on separating ribosomes based on their association with mRNA (polysomes) using fractionation on a density gradient. Since actively translating and stalled ribosomes have the same molecular weight, this method can't differentiate between them.
- RiboLaceTM is Immagina's proprietary technique that selects ribosomes based on their conformational state. By relying on a probe that is specific for ribosomes in the open state, it is possible to specifically isolate and analyze ribosomes that are actively translating mRNA.
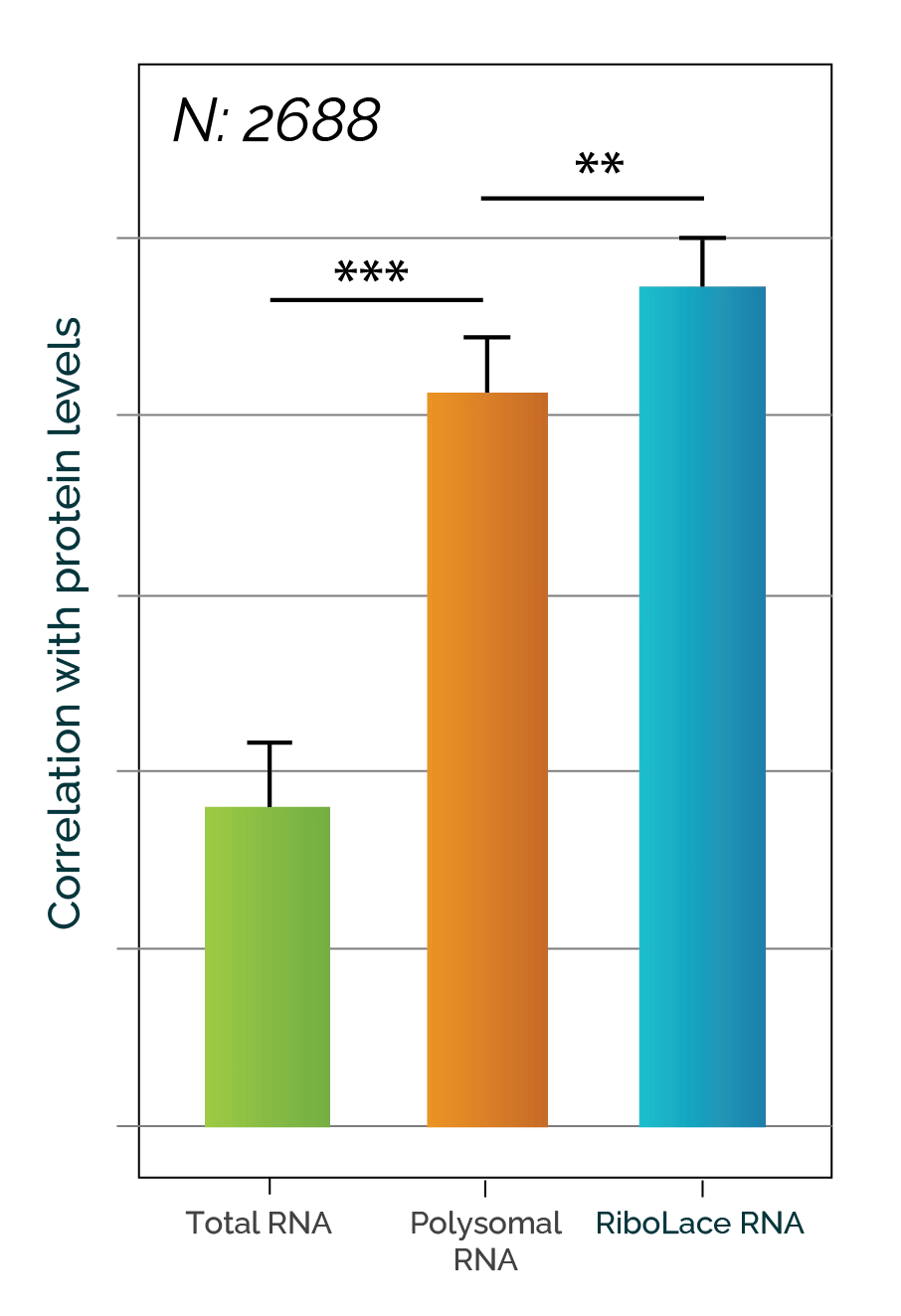
Advantages
- Bypasses the issue of stalled ribosomes
- Provides a more accurate picture of the "active" translation population
- Ensures better correlation with proteomics
- Allows for more robust and affordable experimental workflows
Delivery time: 10 to 12 weeks
Service description and outcomes
- Pre-service consultation: experimental design and BioIT analyses
- Input material: flash-frozen cell pallets or tissues
- RPF pulldown and library preparation
- NGS sequencing
- Bioinformatic analysis
Technical applications
- Studying translational control
- Identification of new mechanisms of translational regulation
- Locating translational start sites and alternative reading frames
- Quantifying translational efficiency

