Technology
LaceSeqTM stands out as a highly effective, accurate, and user-friendly NGS library preparation method tailored for sequencing RNA fragments cleaved by exogenous nucleases, notably in techniques like RiboSeq (ribosome profiling) and CLIP (cross-linking and immunoprecipitation)1. Offering unparalleled clustering efficiency and design precision, LaceSeqTM guarantees:
- High yield of sequencing reads/output
- Accurate identification and trimming of both 3’ and 5’ ends of any RNA fragment
- Minimized rRNA contamination
- Rapid processing time
- Mitigated biases
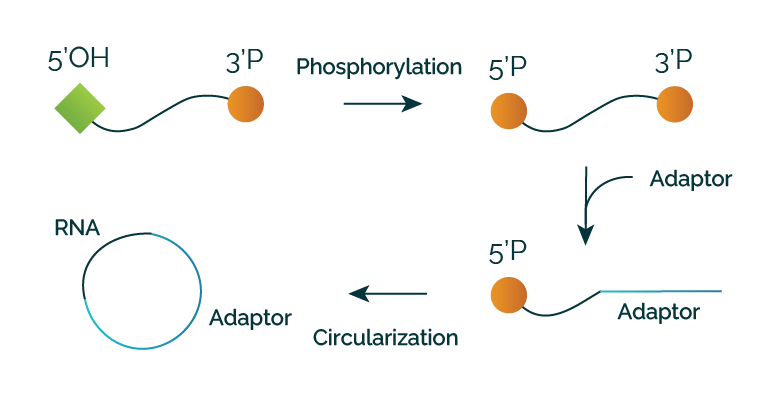
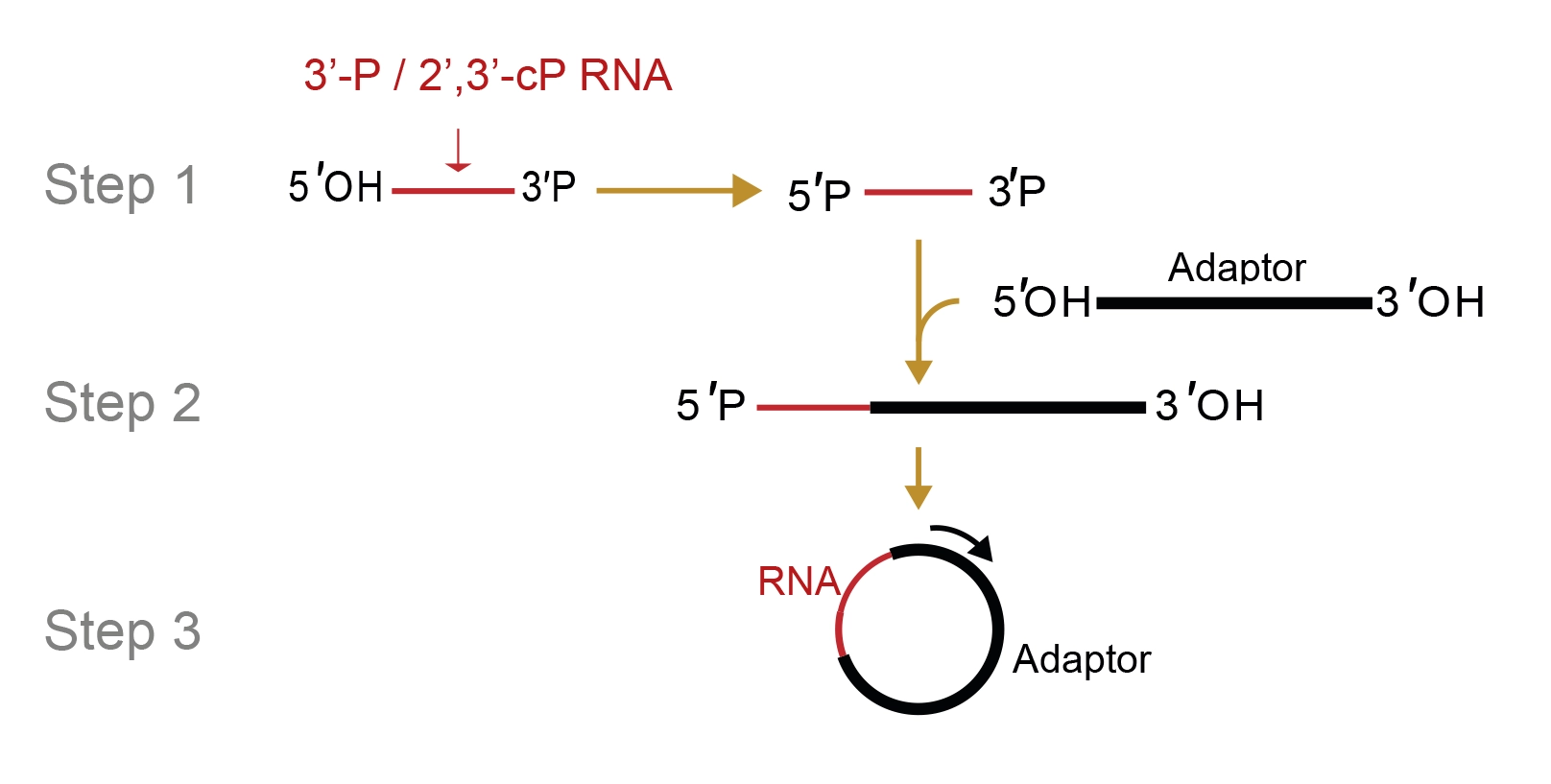
Moreover, this versatile library preparation protocol is well-suited for investigating RNA footprints across various ribosome purification methods or RNA-binding protein (RBP) separation techniques. Typically, commonly used nucleases (e.g. RNase I) result in a phosphate remaining at the 3’ end (3'P) of the cleaved RNA fragments. While traditional methods involve removing this phosphate, LaceSeqTM capitalizes on it to selectively target the digested products, facilitating their preferential integration into the library. Following the initial ligation of the fragment of interest with an RNA-based adaptor modified to increase stability (RNA-drugs design), a secondary, more conventional circularization takes place to minimize RNA contaminants and dimerization products during cDNA synthesis. This kit is exclusively optimized to work with exogenous nucleases. Unique Molecular Identifiers (UMIs) allow the identification of PCR duplication products. Illumina adapters and Unique Dual Indexes (UDIs) for multiplexing are also available. LaceSeqTM can be used as a standalone technology or in combination with RiboLace. It is also available in Oxford Nanopore-compatible version as service only. Immagina BioTechnology s.r.l. exclusively owns and holds full intellectual property rights for LaceSeqTM technology.
Notably, for the profiling of endogenous RNA fragments, Immagina has pioneered DOORs, a cutting-edge technology integrating independent sample preparation, an innovative enzyme for 3’P ligation, and specialized linkers combined with an AI-driven bioinformatic pipeline (P-ROC). DOORs is available for clinical milestone-based projects.
1 -Del Piano, A. et al. Phospho-RNA sequencing with circAID-p-seq. Nucleic Acids Res 50, e23–e23 (2022).
Applications
- NGS-Illumina library of small RNAs
- Ribosome profiling
- Phospho RNA-Seq