Ribosome Profiling
Bridging the gap between mRNA and protein synthesis
Quantify Translation Efficiency - correlate RNA-Seq and proteomics data
Ribo-Seq is a powerful tool for dissecting translational regulation. Traditionally, studies focused on mRNA abundance, but Ribo-Seq provides quantitative insights into ribosome occupancy on transcripts, directly revealing translation efficiency. This allows researchers to pinpoint bottlenecks beyond transcription that might impact protein production.
How it works
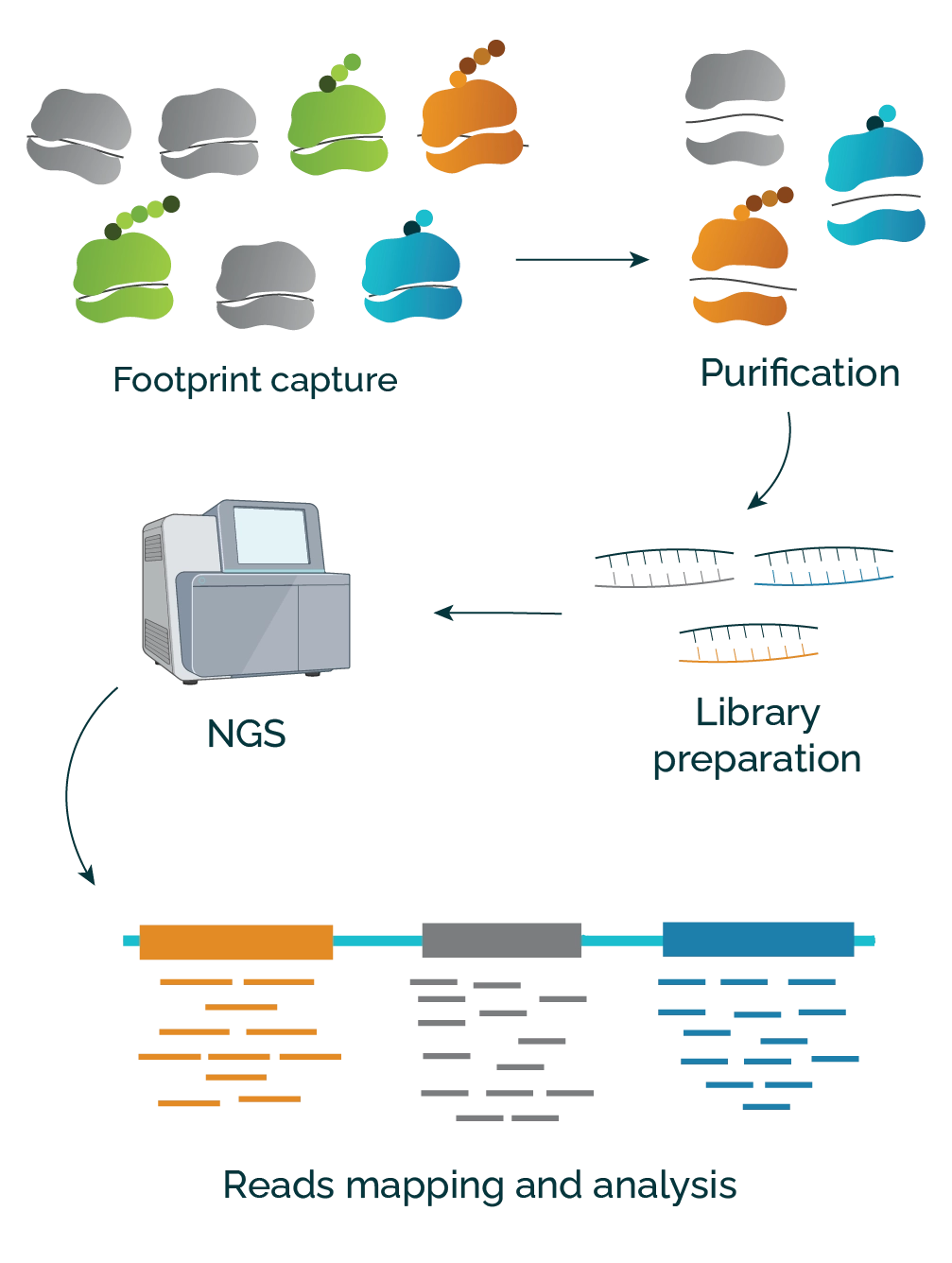
- Footprint Capture: Cells are lysed, then an RNAse digests any RNA not shielded by ribosomes. Ribosome-protected fragments (RPFs) remain, acting as footprints that mark the actively translated regions on messenger RNA.
- Isolation and Sequencing: The RPFs are separated and purified. They are then converted into a library that is suitable for next-generation sequencing (NGS).
- Reads Mapping and Analysis: After NGS, the resulting sequenced fragments (reads) are aligned with a reference genome, and the number of reads that are mapped to each region of the mRNA is counted. More reads indicate a higher concentration of ribosomes, signifying a more actively translated section of the mRNA.
Technical applications
- Investigating translational control
- Identification of new mechanisms of translational regulation
- Locating translational start sites and alternative reading frames
- Quantifying translational efficiency
Delivery time: 10 to 12 weeks
Service description and outcomes
- Pre-service consultation: experimental design and BioIT analyses
- Input material: flash-frozen cell pallets or tissues
- RPF pulldown and library preparation
- NGS sequencing
- Bioinformatic analysis

