Category
RiboSeq category
Applications
LACEseq (12rxns)
LaceSeq streamlines and simplifies library preparation for RiboSeq experiments thanks to:
- Selective 3' End Ligation: LaceSeq utilizes the phosphate group left behind by nucleases used to generate ribosome protected fragments (RPFs). While standard methods typically remove this tag, LaceSeq capitalizes on it to specifically target RPFs for library inclusion.
- Efficient Ligation: Our two-step ligation process preferentially incorporates RPFs while minimizing RNA contaminants, ensuring a cleaner library.
- Optimized Linker: The LaceSeq linker is designed for seamless integration with downstream NGS sequencing and data analysis.
- UMI/UDI: It includes Unique Molecular Identifiers (UMIs) to help identify and remove PCR duplicates, and Illumina adapters - Unique Dual Indexes (iUDIs) for multiplexing are added after the PCR amplification step (iUDIs are sold separately).
- Rapid Library Preparation: The entire process is designed for efficiency, allowing for a quick turnaround.
(Note: The LsceSeq kit does not include iUDIs. Immagina offers four sets of iUDIs , sold separately.)
LaceSeq streamlines and simplifies library preparation for RiboSeq experiments thanks to:
- Selective 3' End Ligation: LaceSeq utilizes the phosphate group left behind by nucleases used to generate ribosome protected fragments (RPFs). While standard methods typically remove this tag, LaceSeq capitalizes on it to specifically target RPFs for library inclusion.
- Efficient Ligation: Our two-step ligation process preferentially incorporates RPFs while minimizing RNA contaminants, ensuring a cleaner library.
- Optimized Linker: The LaceSeq linker is designed for seamless integration with downstream NGS sequencing and data analysis.
- UMI/UDI: It includes Unique Molecular Identifiers (UMIs) to help identify and remove PCR duplicates, and Illumina adapters - Unique Dual Indexes (iUDIs) for multiplexing are added after the PCR amplification step (iUDIs are sold separately).
- Rapid Library Preparation: The entire process is designed for efficiency, allowing for a quick turnaround.
(Note: The LsceSeq kit does not include iUDIs. Immagina offers four sets of iUDIs , sold separately.)
The LACEseq kit creates high-quality NGS libraries from small RNAs with 3' phosphate tags in just one day. Ideal for libraries made from Ribosome Protected Fragments (RPFs) isolated with RiboLace kits or other methods. Compatible with Illumina sequencing platforms.

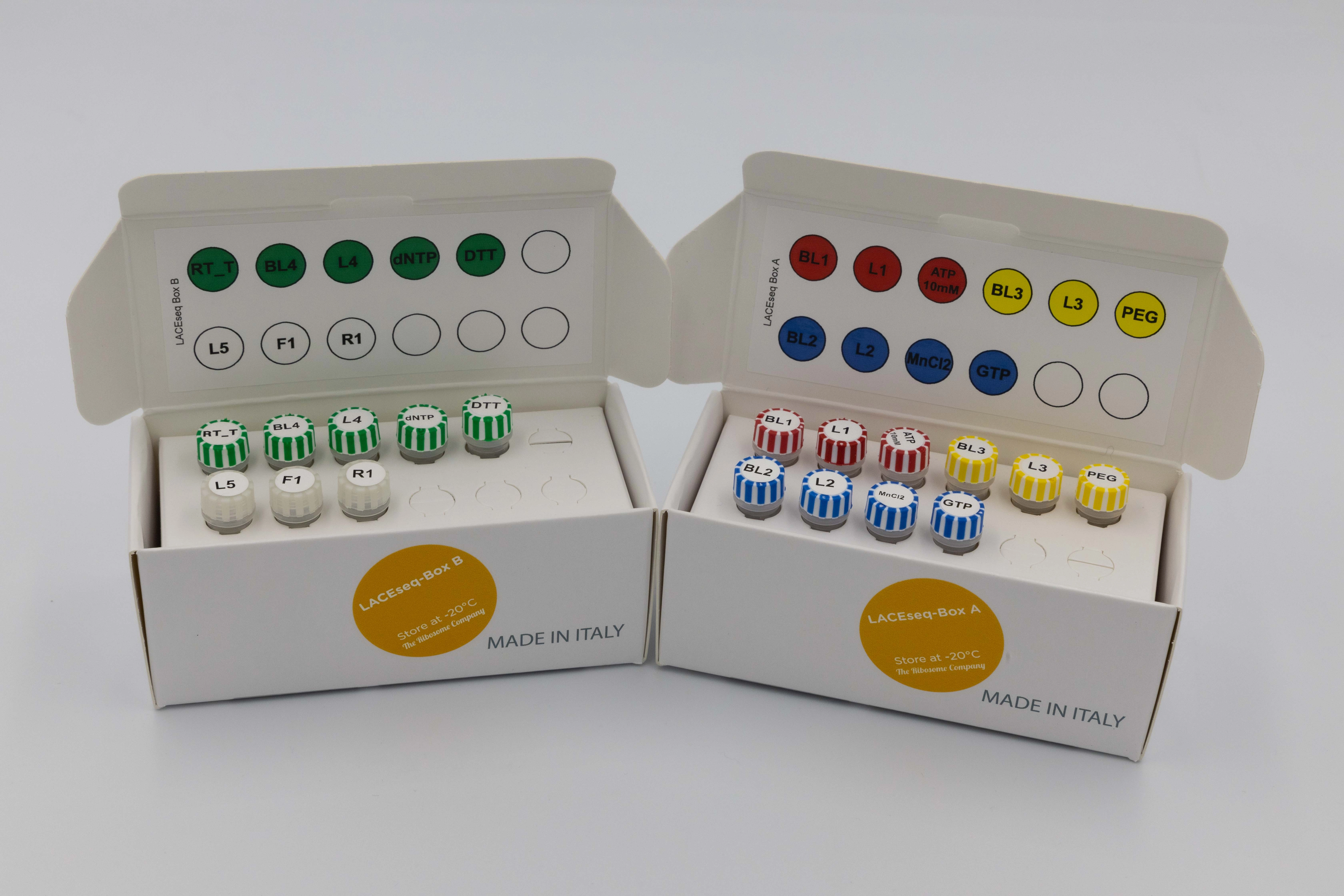

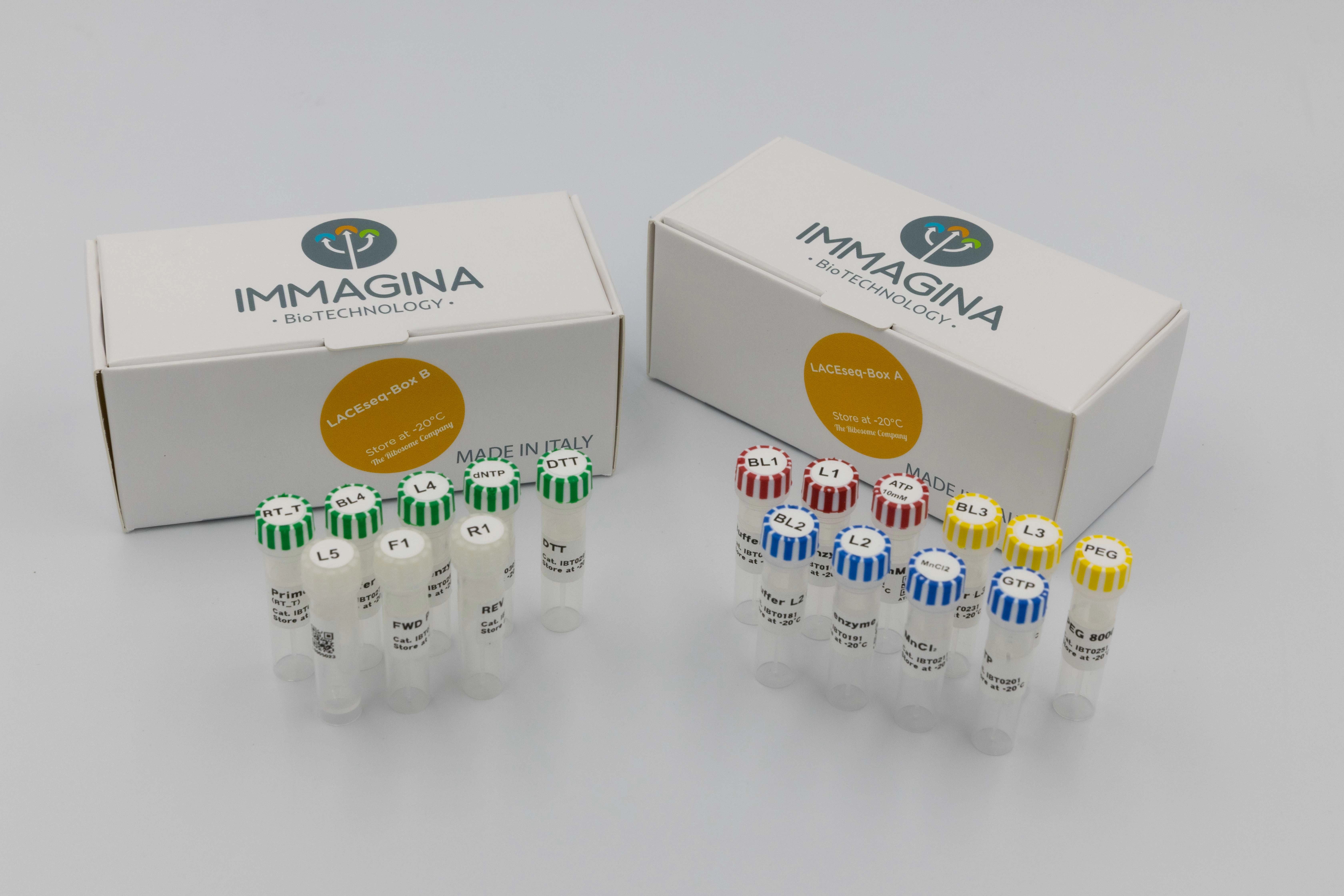












Shipping information
Please note that LACEseq kit will be shipped in dry ice. The shipping is usually scheduled within 1 week from your order. We pride ourselves on rapid and thorough support to ensure your success with our new technology. Please contact us with any questions.
Documentation & research papers